flatbug: A General Insect Detection and Segmentation Model
Motivation
Traditional methods, which often involve manually identifying and counting specimens, are not only labor-intensive but also slow and impractical for large-scale monitoring. With this in mind I, along with my co-author colleagues, have been working hard towards creating and documenting a general tool (flatbug) for insect (and other arthropod) detection and segmentation in images. Finally, we managed to finish our experiments and the accompanying paper entitled “A General Method for Detection and Segmentation of Terrestrial Arthropods in Images” which is now available as a preprint!
The core idea behind flatbug is to achieve consistent and reliable detection and extraction of individual insects across varied imaging conditions and sizes. By integrating YOLOv8 with a robust and diverse training dataset and a novel hyper-inference tiling strategy, flatbug can handle both many different types of images, as well as different organism sizes and densities efficiently.

flatbug, demonstrating versatility across imaging conditions. Each panel displays the predictions of flatbug on a hand-picked ROI—not included in training—for each of the subdatasets within our complete training dataset. The critical improvements achieved with flatbug can be summarized as:
- A diverse segmentation training dataset with a broad taxonomic scope and a wide range of image types and sizes.
- Truly size agnostic and fully segmentation based inference - with an appropriate training regime.
- Appropriate evaluation covering both generalizability, data leakage and domain explainability.
- Easy-to-use APIs for
Python>=3.11and the command line (CLI).
flatbug inference pipeline. Creating a generalized model
To build flatbug, we gathered an extensive and diverse dataset comprising \(113,550\) annotated arthropods from \(6,131\) images collected from both laboratory and field scenarios. This extensive diversity was crucial to ensuring flatbug’s robustness and ability to generalize across previously unseen contexts.
Another key feature that makes flatbug stand apart from similar methods, is our custom-built inference pipeline, which makes flatbug able to work on images of any size.

flatbug's pyramid hyper-inference in action, illustrating effective segmentation across different scales.Because our aim was and is for flatbug to be a truly useful off-the-shelf tool, we were adamant in our validation scheme using both stratified leave-one-out and leave-two-out cross-validation methods. Our results showed that flatbug often only incurs a minor performance reduction (\(\sim 7.1 \%\) relative F1 decrease) when encountering entirely new environments, which indicates that it can indeed be used without any retraining, thus lowering the technical barrier-to-entry significantly.
At the same time we show that flatbug delivers “state-of-the-art” performance with an average F1 score of \(\sim 94 \%\) across the board, demonstrating that flatbug is not only easy to use, but also a powerful model with strong capabilities.
Robust evaluation is inference
In order to really understand the interaction between the models we train, and the data we provide, we set out to design a novel evaluation design and interpretation, based on the aforementioned “leave-two-out” cross-validation. In essence, the design is not so complicated, but it hinges on a somewhat unintuitive insight:
decreases or increases in performance due to the exclusion of a specific pair of datasets from training (on those same datasets, not the whole validation dataset), can be interpreted as a “(dis)similarity” or “distance”.
This meant that by training pairs of leave-two-out cross-validation models for each combination of subdatasets (in practice we only choose 16, since it is quite expensive), we were able to quantify the “closeness”/similarity of our subdatasets, in terms of how they were perceived by the model during training. Having access to such a quantifiable similarity metric, lends itself to the rich body of methods for similarity- or distance-based analysis and interpretation, where some of the simplest are the class of hierarchical agglomerative clustering methods such as UPGMA.
Below the result of said analysis can be found, in which it is clear, that almost all the datasets (due to forming a cluster with a common root below zero) form a synergistic group for learning.
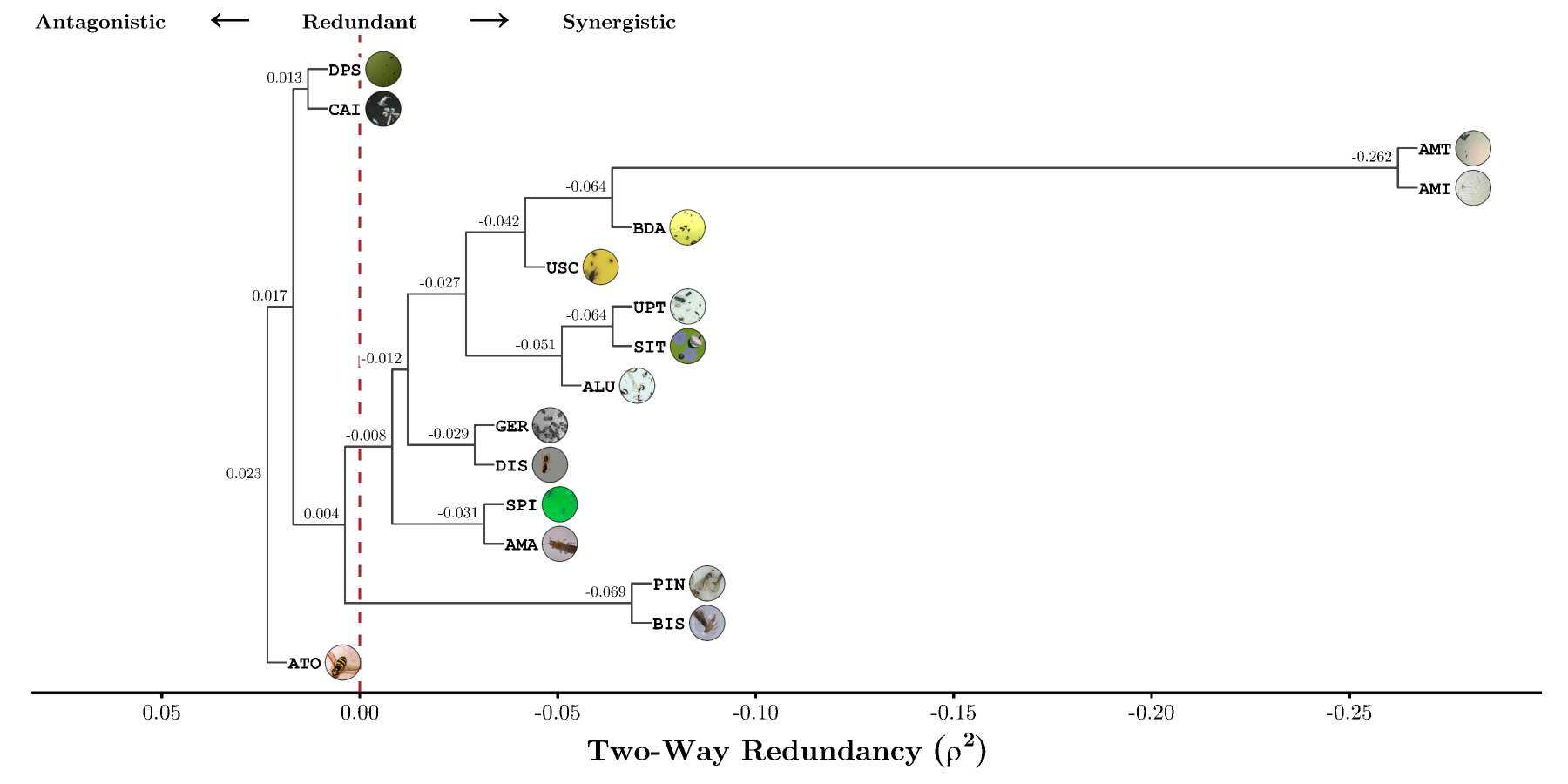
At the same time we can also deduce that the images obtained from citizen science are quite distinct, and do not have such a synergistic relationship, which may not be overly surprising since from a qualitative point of view these images are more diverse and “harder”, in the sense that it requires a broader contextual understanding of the non-standardized conditions with which citizen science images are naturally taken within (extremely varying image quality, viewing angles, background compositions, etc.) to accurately distinguish subject (arthropods) from noise (leaves, flowers, skin, clothing).
Making flatbug accessible
Finally, because I (and my colleagues) believe transparency is a cornerstone in research and a prerequisite for widespread adoption, we have maintained flatbug as a fully open-source and reproducible software and data package. You can find the comprehensive Python package, documentation, tutorials and demo on GitHub, and our rich dataset is publicly available here.
Feel free to explore our work and see how flatbug can revolutionize your monitoring and research projects:
Technical prerequisites
- A consumer grade NVIDA GPU (>2GB VRAM for the smallest model, and >12 GB VRAM for the largest model)
Python>=3.11andPyTorch- An internet connection (for installing the package and downloading models)
Acknowledgments
Thanks all my brilliant co-authors for their contributions: Quentin Geissmann, Guillaume Mougeot, Toke Thomas Høye, Juli Carrillo, Jamie Alison, Daphne Chevalier, Kim Bjerge, Nisa Chavez Molina, and Song-Quan Ong, as well as everyone who helped with data collection, annotations, and testing.