Qualifying Exam: An update on my PhD project and plans
When you’re having fun…
Two out of four years in my PhD have already passed — I guess time does really fly when you’re “having fun”! All jokes aside, these two years have been really great, although sometimes also quite tough, but always always engaging and interesting.
Some reflections
A PhD might not be for everyone — the uncertainty, haphazard schedule, travelling and all considered — but at least in my case it allows an attempt at satisfying my unrelenting curiosity by giving me the freedom to come up with an idea and put my head down under until something or another clicks. I might have been a bit stressed, when I was rushing to finish the ML pipeline before XPRIZE last spring, but at the end of the day the learning experience with building such a pipeline was immense, and the experience of being part of the XPRIZE Rainforest finals in Manaus was hectic and crazy, exhausting and overwhelming, but it was also extremely fun and exciting.
I think that’s a general lesson I’ve learned during these first two years — or at least cemented to a much higher degree than they were in my mind before — things that seem impossible are almost never that, they are hard, sometimes even extremely hard, but if I really try I will succeed and it will be both much more fun, but also satisfying than I thought. So I will try to remember that, as I am sure that there will be plenty more where that comes from in the next two years, but I look forward to it!
A teaser
Below are some hand-picked excerpts from my qualifying exam — that I passed! 🥳🎉
(If you’re intrigued my full progress report contains a lot more details and is available at the bottom of this page)
Major projects in the first half of my PhD
- Detection & segmentation (
flatbug)
Scale- and size-agnostic inference with mask-level outputs; trained on \(113,550\) annotated arthropods across \(23\) subdatasets and \(6,131\) images. Generalizes with an average relative F1 drop of \(\approx 7.1\%\) under stratified leave-one-out; short fine-tuning largely removes penalties.
flatbug's pyramid hyper-inference in action, illustrating effective segmentation across different scales. - Real-world deployment (
XPRIZE Rainforest/ETH BiodivX)
End-to-end laptop pipeline (detection → tracking → hierarchical classification) processed \(\sim 30,000\) frames and yielded \(685,611\) detections in one night, enabling rapid activity curves and richness summaries.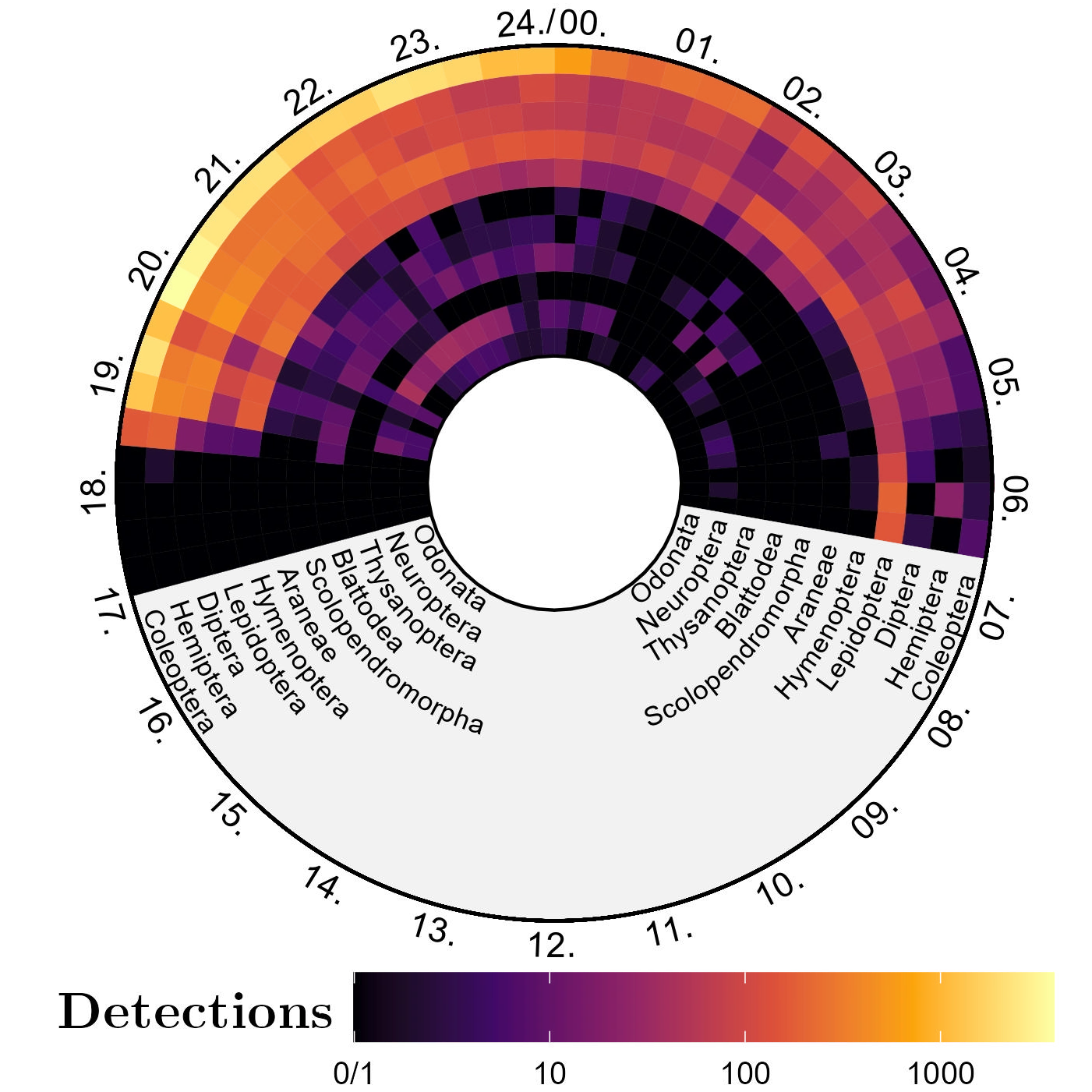
Order-level activity curves in 20-minute bins over a 24-hour axis --- like a clock! --- from one of the ETH BiodivXcanopy camera light trap/rafts. Each concentric circle corresponds to a specific arthropod order (labelled on both ends). - Hierarchical classification
Three model families to be compared — flat, multi-label (multi-head), and hierarchical multi-layer with probability aggregation to genus/family. Evaluation includes top-K plus taxonomic distance/error.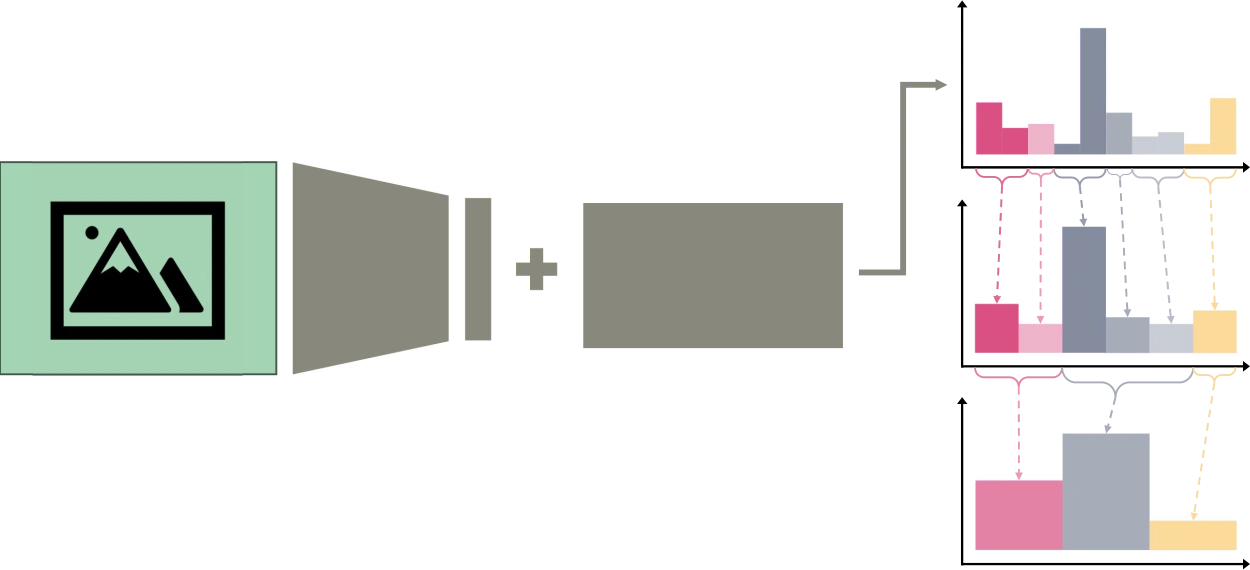
Conceptual diagram for the fixed-hierarchy multi-layer classification model. Each color (pink/grey/yellow) corresponds to one of three conceptual "families" (bottom row), while shades correspond to genera within each (middle row), and bars in the top row are species.
Future plans
Over the next two years, as I finish my PhD project, I plan to continue my research on automated monitoring, with a particular focus on the classification task motivated by two key insights:
- Through my education as a biologist, particular in taxonomy, evolution and bioinformatics, it has always been clear, and only more so as time goes on, that the concept of discrete
"species"is an extremely practical, but at the end of the day arbitrary, delineation which obscures the fact that all organisms are part of the same family, hence all organisms are better viewed as placed in a shared “evolutionary space”. Species then, are just more or less non-intersecting contiguous partitions in this space. In some cases this evolutionary space is very “lumpy” such that clusters of individuals can be separated with high support, however in many cases this is not the case, and instead individuals form a continuum without any natural separation — this is of course also the case for the entire space if we include all individuals that have existed since LUCA. - All attempts at species image classification using computer vision and deep learning so far have at the end of the day relied on species, with almost all methods treating classification as a, well, “classification problem”, when as I just described, the real problem isn’t to tell which species an individual in an image belongs to, but rather where in the evolutionary space the individual is located — seemingly a “regresssion problem”?!
I am aware that some groups have used contrastive learning approaches — e.g. BioCLIP — however the contrastive space is based on human language descriptions of organisms, in particular using "the Linnean Taxonomy"; i.e. using discrete species!
This naturally brings up the question:
Can we change the training objective for species image classification deep learning models from classification to mapping — in other words where in the evolutionary landscape does the imaged individual belong?
And the hypothesis:
If species is an arbitrary concept, and models are trained to classify individuals as if they are instances of discrete entities, when they are not, training models for a different objective, one which is not rooted in arbitrary delineations, should increase stability, “true” learning and generalization.